The increasingly frequent use of dermoscopy makes us think about the possibility of transfer of microorganisms, through the dermatoscope, between doctor and patients.
ObjectivesTo identify the most frequent gram-positive cocci in dermatoscopes and smartphone adapters, as well as the resistance profile, and to evaluate the factors associated with a higher risk of bacterial contamination of the dermatoscopes.
MethodsA cross-sectional study was carried out with 118 dermatologists from Porto Alegre/Brazil between September 2017 and July 2018. Gram-positive cocci were identified by MALDI-TOF MS and habits of use of the dermatoscope were evaluated through an anonymous questionnaire.
ResultsOf the dermatoscopes analysed, 46.6% had growth of gram-positive cocci on the lens and 37.3% on the on/off button. The microorganisms most frequently found were S. epidermidis, S. hominis and S. warneri. Attending a hospital, using the dermatoscope at the hospital, with inpatients and in the intensive care unit were significantly associated with colonisation by gram-positive cocci. The highest resistance rates were observed for penicillin, erythromycin and sulfamethoxazole-trimethoprim.
Study limitationsThe non-search of gram-negative bacilli, fungi and viruses. Moreover, the small number of adapters did not make it possible to better define if the frequency differences were statistically significant.
ConclusionCoagulase-negative staphylococci were frequently identified. S. aureus was detected only on the lens.
Dermoscopy is an excellent diagnostic tool in the daily practice of the dermatologist. In recent years, smartphone adapters have been used to photograph skin lesions, such as melanocytic nevi, and allow their follow-up, as well as facilitate case discussions among dermatologists.
These technological innovations have enabled health professionals to quickly access a wider range of information: using smartphones and tablets it is easier to search for articles, rapidly access relevant topics in books and applications, discuss cases in groups with experts, and participate in teaching future health professionals.1,2 However, the indiscriminate use of such objects provides a new challenge: the possible transference of microorganisms, with or without pathogenic potential, from these devices to the hands of professionals, or vice versa, or person to person transference. For example, patients with nasal colonization by Staphylococcus aureus are 2–9 times more likely to have S. aureus infection.3 The most frequently identified pathogens associated with healthcare-associated infections were Coagulase-Negative Staphylococci (CoNS) (15%), S. aureus (15%), Enterococcus sp. (12%), Candida sp. (11%), followed by several gram-negative bacilli.4 These healthcare-related infections represent a major challenge for the health system and are associated with significant costs, morbidity, and mortality. At any one time, up to 7% of patients in developed and 10% in developing countries will acquire at least one health care-associated infection.5
Few studies have evaluated the contamination of microorganisms in dermatoscopes and there are no studies on smartphone adapters. A study in Switzerland analysed the bacterial presence on the lenses of dermatoscopes belonging to 10 dermatologists (n=10) involved in the care of patients from the dermatology outpatient clinics of two Swiss hospitals. Of the 112 swabs taken, 65% showed the growth of non-pathogenic bacteria (including all the CoNS, Streptococcus α and γ-haemolytic, Corynebacterium, Bacillus and Lactobacillus). Methicillin-Susceptible Staphylococcus Aureus (MSSA) was found on three occasions (on the three swabs, for dermoscopy, immersion oil had been used on the apparatus rather than isopropyl alcohol).6 In Austria, researchers studied the spectrum of microorganisms in 4 dermatoscopes (lens and body) used in the department of dermatology at a Vienna hospital after dermoscopy in 39 patients and found S. epidermidis in 74% of the devices and S. aureus in 7%. In the United Kingdom, Chattopadhyay et al. (2014) evaluated bacterial growth on 9 dermatoscope lenses on 60 occasions (30 before dermoscopy and 30 after). An alcohol-gel containing 70% ethanol was used as the immersion liquid. The authors found Methicillin-Resistant S. Aureus (MRSA) in 10% of the experiments (all from swabs obtained after dermoscopy).7
The present researchers have sought to identify the gram-positive cocci most commonly found on dermatoscopes and smartphone adapters and to assess the factors associated with the increased risk of bacterial contamination.
MethodsBetween September 2017 and July 2018, we conducted a cross-sectional study among dermatologists who attended at a dermatology outpatient clinic of a public hospital and in private practices. The doctors answered an anonymous questionnaire containing demographic information and about the habits of use of the dermatoscope, and provided their devices for bacteriological analysis via the swab technique. Physicians who did not wish to fill out the questionnaire or provide their dermatoscopes or cell phone adapters for swabbing were excluded.
Samples were collected from two or three previously defined sites on the dermatoscopes: the lens, the on/off button and the outside of the smartphone adapter (for professionals who used that device). The swabs were sealed, labelled and forwarded for analysis. In the laboratory, the swabs were placed in tubes containing BHI enriched medium (brain-heart infusion) (Sigma Aldrich, Merck, Germany) and left in the oven at 35±2°C for 24h. In the presence of turbidity (in which case the test is considered positive, i.e. there was bacterial growth), the broths were seeded in 5% sheep-blood agar plates (Biomeriéux, Marcy L’Etoile, France) using a sterile loop. The plates were then returned to the oven at 35±2°C for 24h. The plates showing bacterial growth were placed in skimmed milk and frozen for later identification. Samples that clouded in the BHI broth were subsequently thawed and, again, seeded on blood agar plates for identification by MALDI-TOF MS. In this study, we used the Bruker Daltonics platform (microflex LT; Bruker Daltonik GmbH, Bremen, Germany).
To test for susceptibility, after 24h in culture with sheep-blood agar, the colonies were incubated at 35±2°C for 18–24h and tested for the following antibiotics: penicillin (10U), cefoxitin (30μg), erythromycin (15μg), clindamycin (2μg), levofloxacin (5μg), sulfamethoxazole–trimethoprim (1.25–23.75μg), linezolid (30μg), tetracycline (30μg), gentamicin (10μg) and rifampicin (5μg). The plates were analysed according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI, 2018).8 According to the diameter of the inhibition halo, the samples were classified as sensitive, intermediate resistance or resistant. S. aureus ATCC 25923 was used to control the quality of the antibiotic discs, according to standard disc-diffusion test procedures.
The research was approved by the Research Ethics Committee of the Santa Casa de Misericórdia de Porto Alegre (protocol n° 9396017.5.0000.5335), Universidade Federal de Ciências da Saúde (protocol n° 69396017.5.0001.5345) and Secretaria Estadual de Saúde do Rio Grande do Sul (protocol n° 9396017.5.3002.5312). All the participants included in the study signed an Informed Consent Term.
The data was entered into the Excel program and then exported to the SPSS v. 20.0 for statistical analysis. Qualitative variables were described by frequency and percentages. Symmetrically distributed quantitative variables were described using the mean and standard deviation, while for those with an asymmetric distribution; the median and the interquartile range were used. Categorical variables were compared using the Chi-square test or Fisher's exact test. The quantitative variables were compared using Mann-Whitney test or Student t-Test. A significance level of 5% was considered for the established comparisons.
Regarding the sample size, with approximately 59 dermatologists per group, we were able to detect a difference of 20 percentage points in the frequency of bacterial colonization. We considered a baseline colonization value of 5% (in the cited literature, the value ranges from 2.7% to 10%), a power of 80% and a significance of 5%.
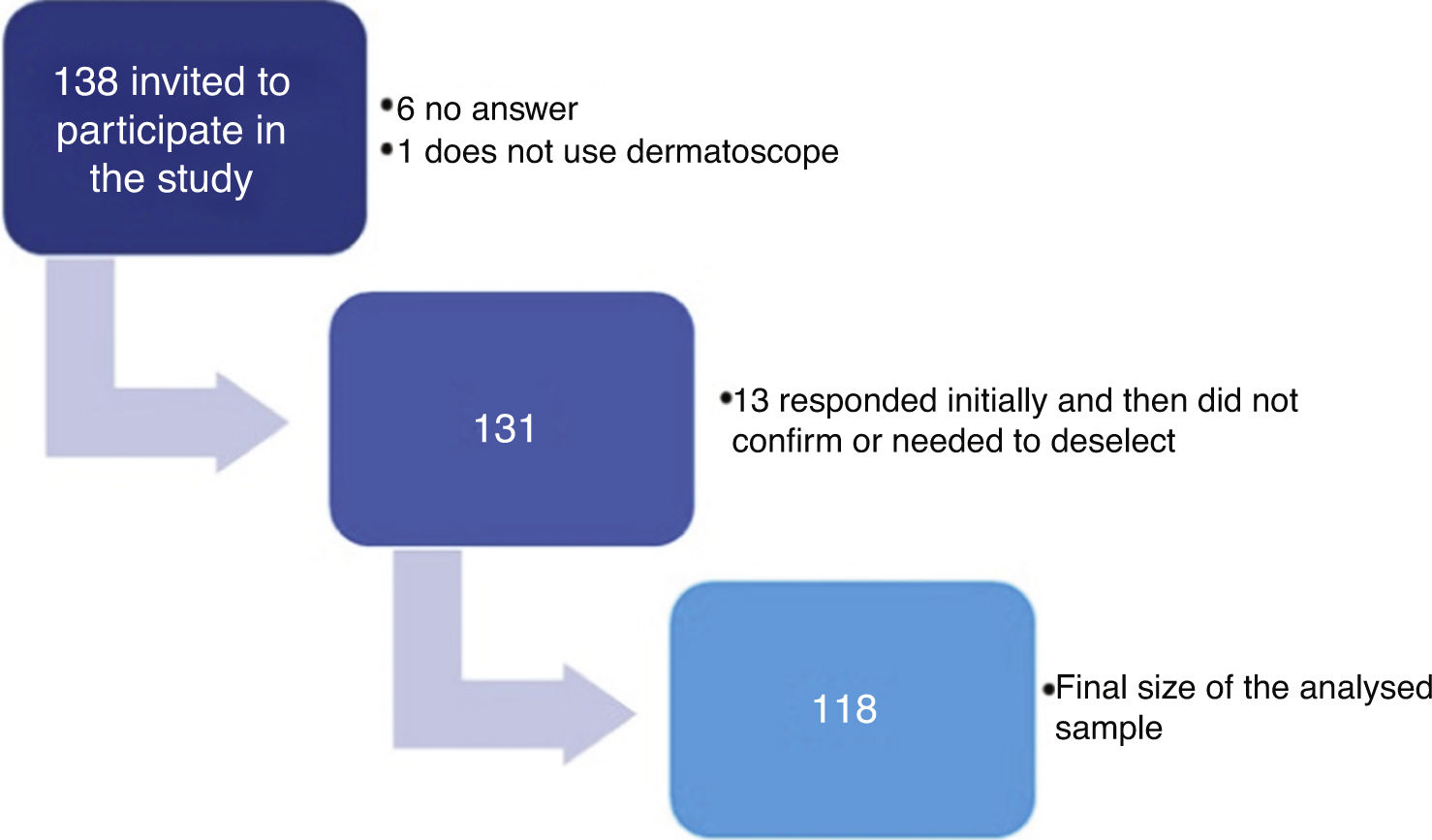
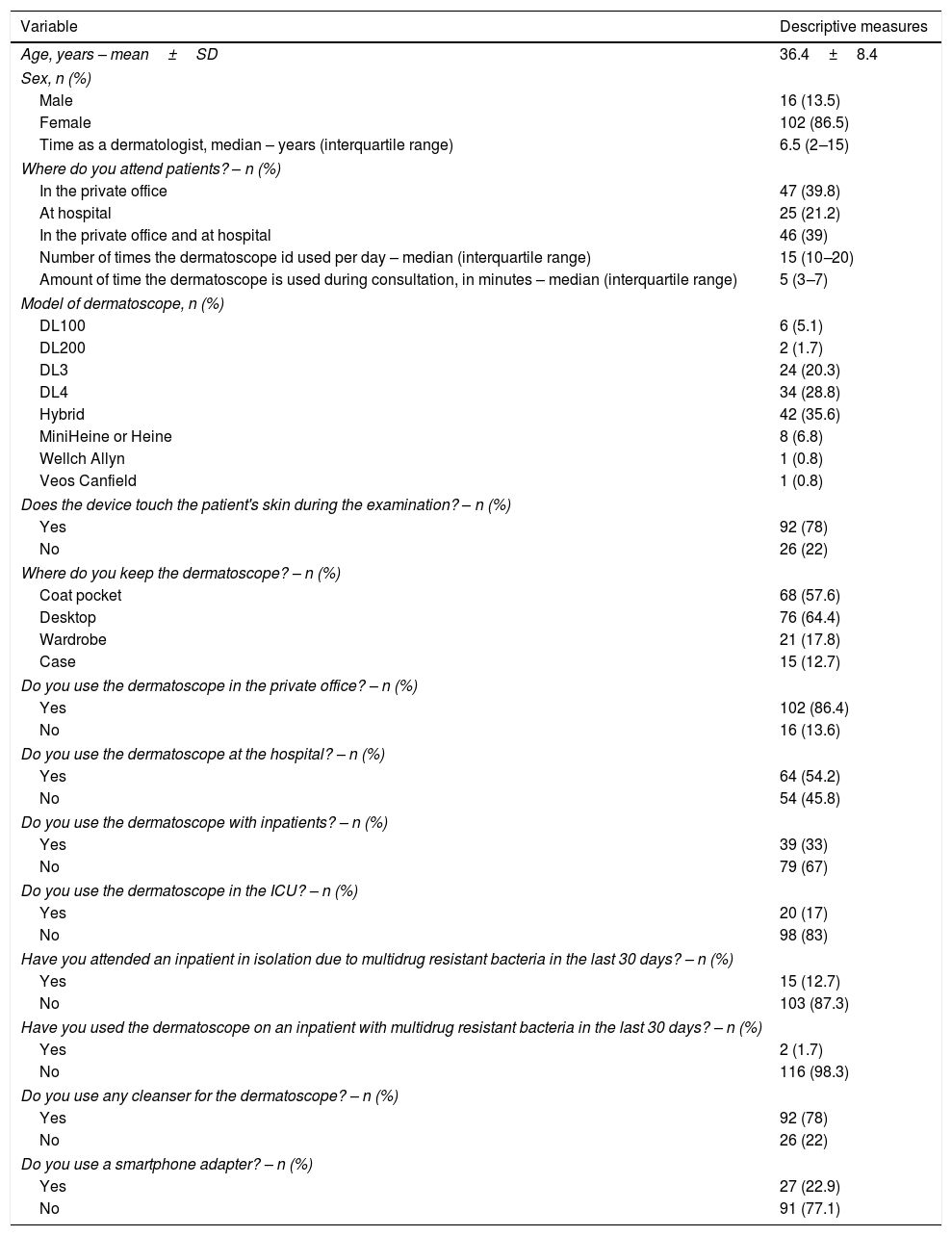
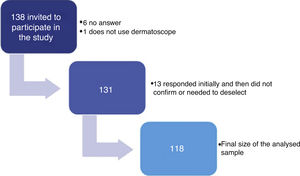
ResultsA total of 138 dermatologists were invited to participate in the study (Fig. 1). The characteristics of the 118 dermatologists whose devices were analysed are described in table 1.
Characteristics of the dermatologists in the sample and the use of the dermatoscope.
| Variable | Descriptive measures |
|---|---|
| Age, years – mean±SD | 36.4±8.4 |
| Sex, n (%) | |
| Male | 16 (13.5) |
| Female | 102 (86.5) |
| Time as a dermatologist, median – years (interquartile range) | 6.5 (2–15) |
| Where do you attend patients? – n (%) | |
| In the private office | 47 (39.8) |
| At hospital | 25 (21.2) |
| In the private office and at hospital | 46 (39) |
| Number of times the dermatoscope id used per day – median (interquartile range) | 15 (10–20) |
| Amount of time the dermatoscope is used during consultation, in minutes – median (interquartile range) | 5 (3–7) |
| Model of dermatoscope, n (%) | |
| DL100 | 6 (5.1) |
| DL200 | 2 (1.7) |
| DL3 | 24 (20.3) |
| DL4 | 34 (28.8) |
| Hybrid | 42 (35.6) |
| MiniHeine or Heine | 8 (6.8) |
| Wellch Allyn | 1 (0.8) |
| Veos Canfield | 1 (0.8) |
| Does the device touch the patient's skin during the examination? – n (%) | |
| Yes | 92 (78) |
| No | 26 (22) |
| Where do you keep the dermatoscope? – n (%) | |
| Coat pocket | 68 (57.6) |
| Desktop | 76 (64.4) |
| Wardrobe | 21 (17.8) |
| Case | 15 (12.7) |
| Do you use the dermatoscope in the private office? – n (%) | |
| Yes | 102 (86.4) |
| No | 16 (13.6) |
| Do you use the dermatoscope at the hospital? – n (%) | |
| Yes | 64 (54.2) |
| No | 54 (45.8) |
| Do you use the dermatoscope with inpatients? – n (%) | |
| Yes | 39 (33) |
| No | 79 (67) |
| Do you use the dermatoscope in the ICU? – n (%) | |
| Yes | 20 (17) |
| No | 98 (83) |
| Have you attended an inpatient in isolation due to multidrug resistant bacteria in the last 30 days? – n (%) | |
| Yes | 15 (12.7) |
| No | 103 (87.3) |
| Have you used the dermatoscope on an inpatient with multidrug resistant bacteria in the last 30 days? – n (%) | |
| Yes | 2 (1.7) |
| No | 116 (98.3) |
| Do you use any cleanser for the dermatoscope? – n (%) | |
| Yes | 92 (78) |
| No | 26 (22) |
| Do you use a smartphone adapter? – n (%) | |
| Yes | 27 (22.9) |
| No | 91 (77.1) |
SD, standard deviation; ICU, intensive care unit.
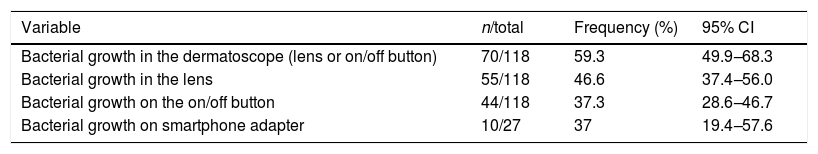
Of the dermatoscopes analysed, 46.6% had gram positive cocci colonies on the lens and 37.3% on the on/off button (Table 2).
Frequency of bacterial colonization by gram-positive cocci in dermatoscopes and smartphone adapters.
| Variable | n/total | Frequency (%) | 95% CI |
|---|---|---|---|
| Bacterial growth in the dermatoscope (lens or on/off button) | 70/118 | 59.3 | 49.9–68.3 |
| Bacterial growth in the lens | 55/118 | 46.6 | 37.4–56.0 |
| Bacterial growth on the on/off button | 44/118 | 37.3 | 28.6–46.7 |
| Bacterial growth on smartphone adapter | 10/27 | 37 | 19.4–57.6 |
CI, confidence interval.
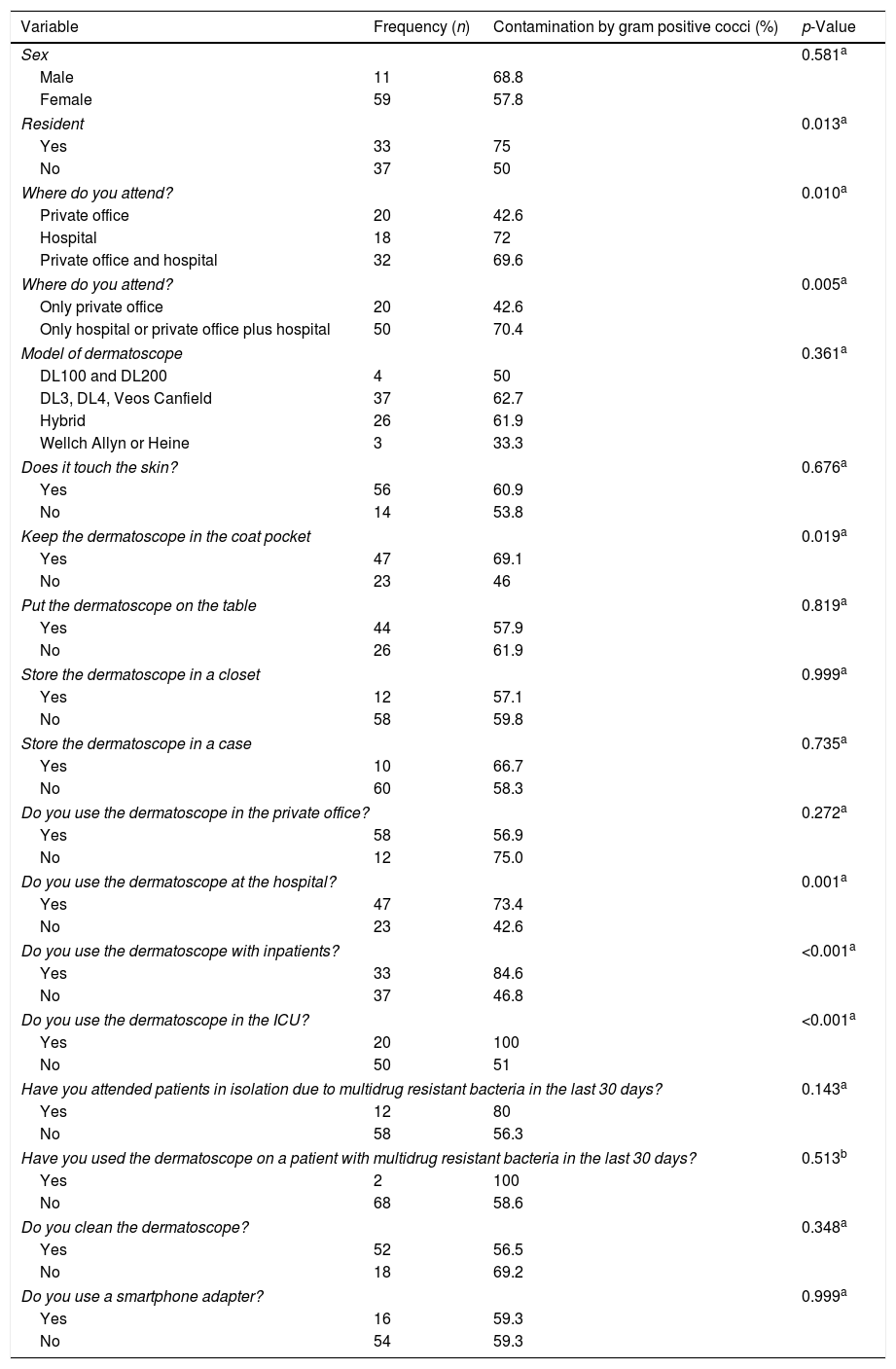
The frequency of gram-positive cocci was higher among males, but the difference was not statistically significant (Table 3). The variables significantly associated with colonisation by gram-positive cocci were: being a resident. Attending a hospital or not attending exclusively in private office; Keeping the dermatoscope in the coat pocket; Using the dermatoscope at the hospital; With inpatients and; In the Intensive Care Unit (p<0.05). Using a smartphone adapter was not associated with dermatoscope contamination.
Categorical variables and their relation to the presence of bacterial contamination on the lens or on the on/off button.
| Variable | Frequency (n) | Contamination by gram positive cocci (%) | p-Value |
|---|---|---|---|
| Sex | 0.581a | ||
| Male | 11 | 68.8 | |
| Female | 59 | 57.8 | |
| Resident | 0.013a | ||
| Yes | 33 | 75 | |
| No | 37 | 50 | |
| Where do you attend? | 0.010a | ||
| Private office | 20 | 42.6 | |
| Hospital | 18 | 72 | |
| Private office and hospital | 32 | 69.6 | |
| Where do you attend? | 0.005a | ||
| Only private office | 20 | 42.6 | |
| Only hospital or private office plus hospital | 50 | 70.4 | |
| Model of dermatoscope | 0.361a | ||
| DL100 and DL200 | 4 | 50 | |
| DL3, DL4, Veos Canfield | 37 | 62.7 | |
| Hybrid | 26 | 61.9 | |
| Wellch Allyn or Heine | 3 | 33.3 | |
| Does it touch the skin? | 0.676a | ||
| Yes | 56 | 60.9 | |
| No | 14 | 53.8 | |
| Keep the dermatoscope in the coat pocket | 0.019a | ||
| Yes | 47 | 69.1 | |
| No | 23 | 46 | |
| Put the dermatoscope on the table | 0.819a | ||
| Yes | 44 | 57.9 | |
| No | 26 | 61.9 | |
| Store the dermatoscope in a closet | 0.999a | ||
| Yes | 12 | 57.1 | |
| No | 58 | 59.8 | |
| Store the dermatoscope in a case | 0.735a | ||
| Yes | 10 | 66.7 | |
| No | 60 | 58.3 | |
| Do you use the dermatoscope in the private office? | 0.272a | ||
| Yes | 58 | 56.9 | |
| No | 12 | 75.0 | |
| Do you use the dermatoscope at the hospital? | 0.001a | ||
| Yes | 47 | 73.4 | |
| No | 23 | 42.6 | |
| Do you use the dermatoscope with inpatients? | <0.001a | ||
| Yes | 33 | 84.6 | |
| No | 37 | 46.8 | |
| Do you use the dermatoscope in the ICU? | <0.001a | ||
| Yes | 20 | 100 | |
| No | 50 | 51 | |
| Have you attended patients in isolation due to multidrug resistant bacteria in the last 30 days? | 0.143a | ||
| Yes | 12 | 80 | |
| No | 58 | 56.3 | |
| Have you used the dermatoscope on a patient with multidrug resistant bacteria in the last 30 days? | 0.513b | ||
| Yes | 2 | 100 | |
| No | 68 | 58.6 | |
| Do you clean the dermatoscope? | 0.348a | ||
| Yes | 52 | 56.5 | |
| No | 18 | 69.2 | |
| Do you use a smartphone adapter? | 0.999a | ||
| Yes | 16 | 59.3 | |
| No | 54 | 59.3 | |
ICU, intensive care unit.
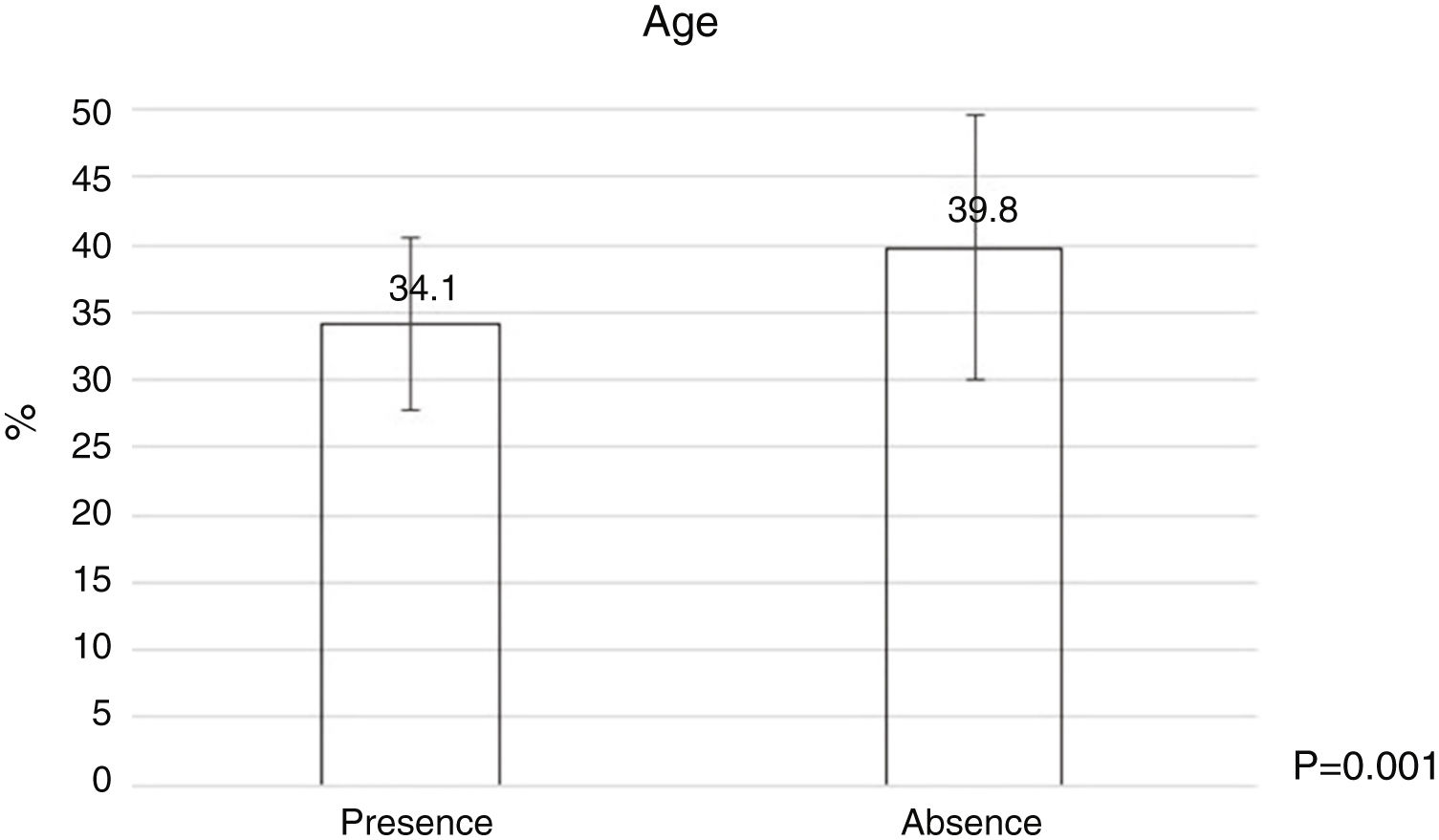
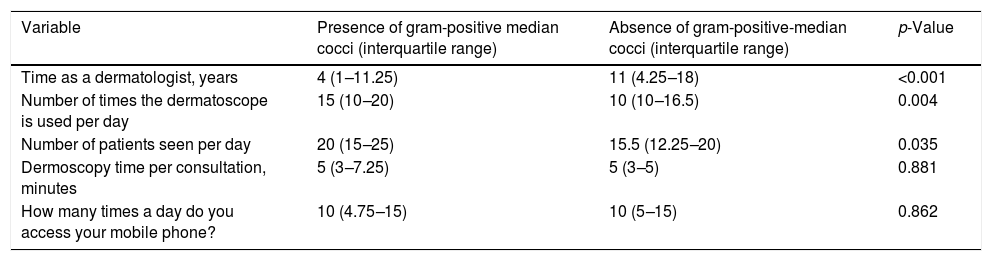
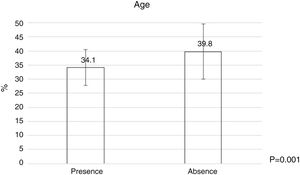
Bacterial contamination was more common among younger dermatologists (Fig. 2) and those wiht less time as dermatologist (Table 4), and a statistically significant relationship was found between the number of patients treated per day and the number of times they used the dermatoscope per day (Table 4).
Numerical variables and their relation to the presence or absence of bacterial contamination on the lens or on the on/off button.
| Variable | Presence of gram-positive median cocci (interquartile range) | Absence of gram-positive-median cocci (interquartile range) | p-Value |
|---|---|---|---|
| Time as a dermatologist, years | 4 (1–11.25) | 11 (4.25–18) | <0.001 |
| Number of times the dermatoscope is used per day | 15 (10–20) | 10 (10–16.5) | 0.004 |
| Number of patients seen per day | 20 (15–25) | 15.5 (12.25–20) | 0.035 |
| Dermoscopy time per consultation, minutes | 5 (3–7.25) | 5 (3–5) | 0.881 |
| How many times a day do you access your mobile phone? | 10 (4.75–15) | 10 (5–15) | 0.862 |
Statistical test used: Mann–Whitney test.
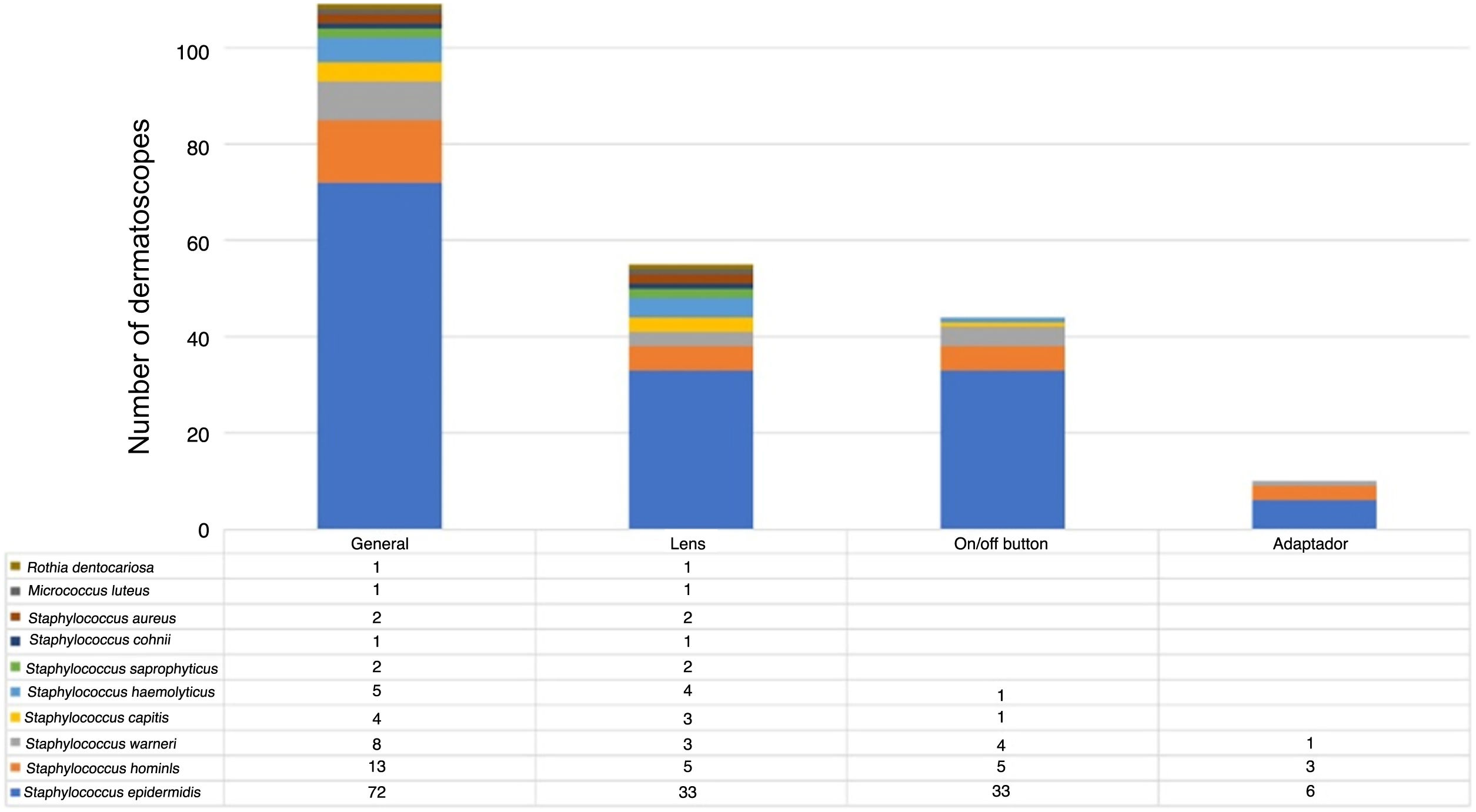
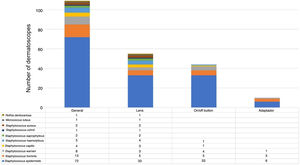
The microorganisms most frequently found were S. epidermidis, S. hominis and S. warneri. S. aureus was only detected on the lens (Fig. 3).
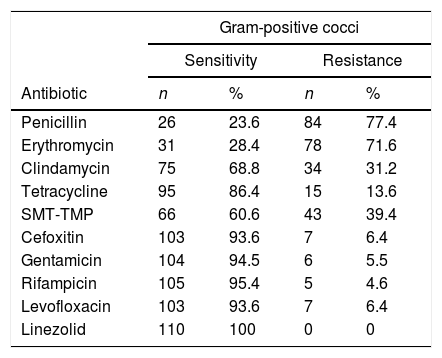
The highest resistance rates among the gram-positive cocci were found to be against penicillin, erythromycin, Sulfamethoxazole-Trimethoprim (SMT-TMP) and clindamycin (Table 5). Cefoxitin resistance was 6.6% and no microorganism was resistant to linezolid.
Antimicrobial resistance profile of gram-positive cocci isolates obtained from dermatoscopes and smartphone adapters.
| Gram-positive cocci | ||||
|---|---|---|---|---|
| Sensitivity | Resistance | |||
| Antibiotic | n | % | n | % |
| Penicillin | 26 | 23.6 | 84 | 77.4 |
| Erythromycin | 31 | 28.4 | 78 | 71.6 |
| Clindamycin | 75 | 68.8 | 34 | 31.2 |
| Tetracycline | 95 | 86.4 | 15 | 13.6 |
| SMT-TMP | 66 | 60.6 | 43 | 39.4 |
| Cefoxitin | 103 | 93.6 | 7 | 6.4 |
| Gentamicin | 104 | 94.5 | 6 | 5.5 |
| Rifampicin | 105 | 95.4 | 5 | 4.6 |
| Levofloxacin | 103 | 93.6 | 7 | 6.4 |
| Linezolid | 110 | 100 | 0 | 0 |
SMT-TMP, sulfamethoxazole-trimethoprim.
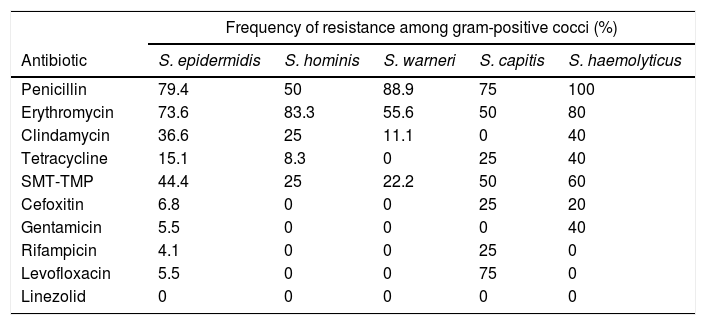
S. epidermidis presented a high rate of resistance to penicillin, erythromycin and SMP-TMP, whereas S. hominis presented greater resistance to erythromycin than penicillin. While S. capitis presented high resistance rates to several antibiotics, no case of resistance to clindamycin and gentamicin was found. The highest rates of resistance to penicillin were found for S. warneri and S. haemolyticus. All isolates of S. haemolyticus were resistant to penicillin and it had the highest frequencies of resistance to clindamycin, tetracycline, SMT-TMP and gentamicin (Table 6).
Antimicrobial resistance profile of the isolates of the most frequent gram-positive cocci obtained from dermatoscopes and smartphone adapters.
| Frequency of resistance among gram-positive cocci (%) | |||||
|---|---|---|---|---|---|
| Antibiotic | S. epidermidis | S. hominis | S. warneri | S. capitis | S. haemolyticus |
| Penicillin | 79.4 | 50 | 88.9 | 75 | 100 |
| Erythromycin | 73.6 | 83.3 | 55.6 | 50 | 80 |
| Clindamycin | 36.6 | 25 | 11.1 | 0 | 40 |
| Tetracycline | 15.1 | 8.3 | 0 | 25 | 40 |
| SMT-TMP | 44.4 | 25 | 22.2 | 50 | 60 |
| Cefoxitin | 6.8 | 0 | 0 | 25 | 20 |
| Gentamicin | 5.5 | 0 | 0 | 0 | 40 |
| Rifampicin | 4.1 | 0 | 0 | 25 | 0 |
| Levofloxacin | 5.5 | 0 | 0 | 75 | 0 |
| Linezolid | 0 | 0 | 0 | 0 | 0 |
SMT-TMP, sulfametoxazol-trimetropim.
The dermatoscopes were mainly colonized by bacteria from the cutaneous microbiota (CoNS), the most frequently found being S. epidermidis, which is accordance with the literature.9,10 This microorganism has become the most common cause of primary bacteraemia and infection of medical devices, such as catheters, particularly in immunocompromised individuals and neonates. In contrast to S. aureus, which is much more virulent and synthesizes an array of toxins and other virulence factors, the main known virulence factor associated with S. epidermidis is its ability to form biofilms and colonize biomaterials. Again, in contrast to S. aureus, which is commonly located in the nasal mucosa, S. epidermidis can be easily transferred to the skin of other individuals through simple contact.11
Both S. hominis, the second most common microorganism found in our study, which is cited in the literature as one of the three CoNS most frequently found in neonatal blood cultures and immunosuppressed patients,12 and S. warneri, the third most common microorganism identified in dermatoscopes in our study, which some articles suggest is the second most frequent CoNS,13,14 have the capacity to form biofilm,15,16 and have been associated with bacteraemia, septicaemia, and endocarditis.16,17
S. capitis rarely causes infection in adults, but a decreased susceptibility to vancomycin has been reported and a clonal population of methicillin-resistant S. capitis with vancomycin heteroresistance has spread among several neonatal ICUs in France and elsewhere.18 Ehlersson et al. evaluated S. capitis isolates from neonatal hemocultures in Sweden and found a 75% cefoxitin and gentamicin resistance rate, only 3% erythromycin resistance, and no case of resistance either to norfloxacin or SMT-TMP.19
We found that the dermatoscope can carry S. aureus. This bacteria colonizes the superficial layer of the skin, survives for a short period of time, and is often acquired by health professionals during direct contact with the patient (colonized or infected), environment, surfaces close to the patient, contaminated products and equipment.20,21 In fact in our study, it was detected precisely on the lens but not on the on-off button or smartphone adapter, places more likely to be related to direct contact with the skin of the health professional.
Hands of health professionals may be persistently colonized by pathogenic microorganisms (such as S. aureus, gram-negative bacilli or yeasts), which in critical areas, such as intensive care units and units with immunocompromised or surgical patients, can play an important role as a cause of infection related to health care.22
In our study, cefoxitin resistance was considered low. Resistance to erythromycin was notably high in S. hominis isolates (83.3%), a fact already mentioned in other studies.23 Szczuka et al. (2016) found an erythromycin resistance rate of 75% in isolates from blood and surgical wounds of hospitalized patients.24 The highest rates of resistance to various antibiotics were seen in S. haemolyticus, as previously reported.23 Recent studies have cited S. haemolyticus as the second CoNS, after S. epidermidis, most frequently isolated from clinical cases, including sepsis patients.25–27
This is the first study in the literature to evaluate antimicrobial resistance of CoNS in dermatoscopes and smartphone adapters. Knowing the resistance pattern of CoNS in dermatoscopes, and on our own skin, is important given that this bacterial group can act as reservoir of antimicrobial resistant genes by horizontal transfer between staphylococcal species. Furthermore, they may be acquired by S. aureus,26,28,29 and subsequently transferred between dermatologists and their patients, especially physicians working in hospital settings where antimicrobial resistance rates are highest.26 According to a cohort of 2518 patients in Israel, identifying the CoNS resistance patterns obtained by blood cultures, even when contaminants, could help predict mortality and correct empirical antibiotic therapy.30
Among the limitations of our study is the non-screening of gram-negative bacteria, fungi, and viruses. Moreover, the small number of adapters meant it we were unable better determine whether the differences in frequency were statistically significant.
ConclusionsWe identified a high frequency of gram-positive cocci on the tested devices. Staphylococcus epidermidis was the most frequently observed, both on the lens, the on/off button and the smartphone adapter. S. aureus was detected only on the lens.
This study concerns the association between the dermatologist and the contamination of dermatoscopes. Professionals should take measures to prevent contamination of their devices and cross-colonization with their patients.
Financial supportCoordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Pesquisa e Desenvolvimento Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Rio Grande do Sul (FAPERGS).
Authors’ contributionsMaurício de Quadros: Statistic analysis; approval of the final version of the manuscript; conception and planning of the study; elaboration and writing of the manuscript; obtaining, analysis, and interpretation of the data; effective participation in research orientation; critical review of the literature; critical review of the manuscript.
Roberto Carlos Freitas Bugs: Approval of the final version of the manuscript; conception and planning of the study; obtaining, analysis, and interpretation of the data; critical review of the literature.
Renata de Oliveira Soares: Approval of the final version of the manuscript; conception and planning of the study; obtaining, analysis, and interpretation of the data; effective participation in research orientation.
Adriana Medianeira Rossato: Approval of the final version of the manuscript; conception and planning of the study; elaboration and writing of the manuscript; obtaining, analysis, and interpretation of the data; effective participation in research orientation; critical review of the literature; critical review of the manuscript.
Lisiane da Luz Rocha: Approval of the final version of the manuscript; conception and planning of the study; elaboration and writing of the manuscript; obtaining, analysis, and interpretation of the data; critical review of the manuscript.
Pedro Alves d’Azevedo: Statistic analysis; approval of the final version of the manuscript; conception and planning of the study; elaboration and writing of the manuscript; obtaining, analysis, and interpretation of the data; effective participation in research orientation; critical review of the literature; critical review of the manuscript.
Conflicts of interestNone declared.
How to cite this article: Quadros M, Bugs RCF, Soares RO, Rossato AM, Rocha LL, d’Azevedo PA. Identifying gram-positive cocci in dermatoscopes and smartphone adapters using MALDI-TOF MS: a cross-sectional study. An Bras Dermatol. 2020;95:298–306.
Study conducted at the Gram-positive Cocci Laboratory, Universidade Federal de Ciências da Saúde de Porto Alegre, Porto Alegre, RS, Brazil.