Background: Behçet disease is a prototypical systemic autoimmune disease, caused by a complex interplay between environmental and genetic factors. The transmembrane immunoglobulin and mucin domain-3 (TIM-3) is a distinct member of the TIM family that is preferentially expressed on Th1 cells and plays a role in Thl-mediated autoimmune or inflammatory diseases, such as Behçet disease.
Objectives: The aim of this study was to test the potential association between TIM-3 gene polymorphisms and Behçet disease.
Methods: Two single-nucleotide polymorphisms of TIM-3 (rs9313439 and rs10515746) were genotyped in 212 patients with Behcet disease and 200 healthy controls. Typing of the polymorphisms was performed using multiplex PCR amplification.
Results: There were no significant differences in allele and genotype frequencies between the Behcet disease patients and controls who were successfully genotyped. Similar results were also found after stratification by gender, age, or clinical features.
Study limitations: Lack of studies on various racial or ethnic groups and small sample size.
Conclusion: This study failed to demonstrate any association between the tested TIM-3 polymorphisms and Behçet disease.
T cells play an important role in the pathogenesis and progression of BD, which is characterized by recurrent oral and genital aphthous ulcers, uveitis, and skin lesions.1 There is also growing evidence of a connection between BD development and genetic variants of immune-related genes.2 Interestingly, some of these variants that may act as predisposing risk factors for BD are T-cell-related molecules (both on the cell surface or internally) and have a close association with T cell activities.3-5
Since BD has been predominantly attributed to be a Th1-driven autoimmune disorder, it seems reasonable to think that variability within Th1 cell-mediated immune response genes may influence disease susceptibility or severity.6 Therefore, some efforts have been made to address this issue and the results have revealed numerous genetic associations between specific single-nucleotide polymorphisms (SNPs) in Th1 related cytokine genes and disease risk.7-9
In recent years, special attention has been given to studies regarding the TIM gene family as a relatively new class of molecules that are involved in the regulation of T cell responses.10 TIM-3, a member of TIM family, is selectively expressed on the surface of differentiated Th1 cells and may play a role in the induction of autoimmune diseases.
The analysis of TIM-3 polymorphisms appears important because the results of different studies indicate an association between the functional variant in TIM-3 and increased risk of autoimmunity.11,12 To date, no study has been conducted to reveal the potential relationship between TIM-3 polymorphisms and susceptibility to BD. Thus, the goal of this study was to evaluate the effects of the two TIM-3 polymorphisms on the risk of BD among an Iranian population.
MethodsPatientsVenous bloods were drawn from 212 patients with BD (134 men, 78 women) in outpatient clinics at teaching hospitals affiliated to Tehran University of Medical Sciences.
The revised international criteria for Behcet’s disease was used for disease diagnosis.13 The mean age of the patients in this study was 38.38 ± 0.72 years (men: 36.99 ± 0.79 years; women: 40.76 ± 1.39 years). The mean duration of BD was 14.65 ± 0.59 years (women: 17.06 ± 1.07 years; men: 13.24 ± 0.66 years).
Peripheral blood samples were also taken from 200 healthy volunteers (143 men, 57 women) referred to the Iranian Blood Transfusion Organization, Tehran, Iran. All controls did not have evidence of BD or other autoimmune diseases and had a mean age of 36.14 ± 0.66 years (men 37.18 ± 0.80 years, women 33.51 ± 0.99 years).
Patients display diverse clinical features encompassing different organ systems to varying degrees. Demographic and clinical characteristics of patients are found in table 1.
Clinical and demographic features of the BD patients and controls
| Features | Patients | Controls |
|---|---|---|
| Number | 212 | 200 |
| Age (years) | 38.38 ± 0.72 | 36.14 ± 0.66 |
| Duration of disease (years) | 14.65 ± 0.59 | |
| Gender (female/male) | 78/134 | 57/143 |
| n/total n (%) | n/total n (%) | |
| Oral aphthosis | 209/212 (98.6) | 0/200(0) |
| Genital aphthosis | 131/212 (61.8) | 0/200(0) |
| Skin lesions | 127/212 (59.9) | |
| Ophthalmic manifestations | 147/212 (69.3) | |
| Joint manifestations | 110/212 (51.9) | |
| Neurological involvement | 25/212 (11.8) | |
| Vascular involvement | 20/212 (9.4) | |
| Other manifestations | 21/212 (9.9) | |
| Pathergy phenomenon | 90/212 (42.5) | |
| Family history of BD | 10/212 (4.7) |
n/total, n (%), number of individuals/total number of patients or controls analyzed; BD, Behcet’s disease.
The proposed experimental protocol was reviewed and approved by the Research Ethics Board at the Tehran University of Medical Sciences. Informed consent was obtained from all participants prior to beginning the research.
SNP selectionThe review of the literature proves that genetic variations of TIM-3 may enhance susceptibility to autoimmune diseases. Nonetheless, there is no evidence regarding the association of TIM-3 polymorphisms and BD. Thus, the present study focused on two SNPs, which were previously investigated in systemic lupus erythematosus and rheumatoid arthritis, two important autoimmune rheumatic diseases.11-14
DNA extractionPeripheral blood was collected in EDTA-treated tubes and DNA was extracted from whole blood using a DNA extraction kit (MBST – Iran) according to the manufacturer’s instructions.
The quality and quantity of DNA were estimated by agarose gel electrophoresis (1.5% w/v agarose gel in 1 × TBE) stained with ethidium bromide) and UV spectrophotometry. Genomic DNA samples were stored at −20 °C until testing.
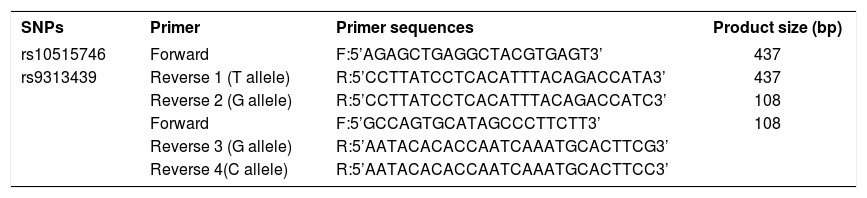
Primer designPCR primers pairs were designed by using OLIGO software (National Biosciences) and purchased from Gene Fanavaran (Tehran, Iran). Two reverse primers were designed for each SNP. The bases at the 3’ end of the reverse primers had variable nucleotide and the remaining sequence being the same. Details of PCR primers used in this study are given in table 2. Bold and underlined letters means different nucleotides between primers.
Primer sequences used in studied polymorphisms
| SNPs | Primer | Primer sequences | Product size (bp) |
|---|---|---|---|
| rs10515746 | Forward | F:5’AGAGCTGAGGCTACGTGAGT3’ | 437 |
| rs9313439 | Reverse 1 (T allele) | R:5’CCTTATCCTCACATTTACAGACCATA3’ | 437 |
| Reverse 2 (G allele) | R:5’CCTTATCCTCACATTTACAGACCATC3’ | 108 | |
| Forward | F:5’GCCAGTGCATAGCCCTTCTT3’ | 108 | |
| Reverse 3 (G allele) | R:5’AATACACACCAATCAAATGCACTTCG3’ | ||
| Reverse 4(C allele) | R:5’AATACACACCAATCAAATGCACTTCC3’ |
SNP, Single Nucleotide Polymorphisms; bp, base pair.
Genotype determination was carried out by multiplex polymerase chain reaction (multiplex-PCR). Two SNPs spanning the TIM-3 gene were used in our association studies, rs9313439 and rs10515746.
PCR amplification was performed in two separate tubes containing respective reverse primers together with a common forward primer for each allele. The reaction mixture contained 12.5μL of Taq DNA Pol 2x Master Mix Red (Ampliqon; Denmark), 0.5 μL of each forward and reverse primers (10 pmoL/μL), 2μL of template DNA (1 ng/μL), and 8.5 μL autoclaved distilled water, making the final volume 25μL. Polymerase chain reaction was performed using on a programmable thermal cycler (Techne Flexigen – Cambridge, United Kingdom).
The amplification conditions were as follows: a pre-denaturation of 94 °C for 5 min, 38 cycles of amplification (95 °C for 1 min, 62 °C for 1 min, and 72 °C for 45s) and a final extension reaction was performed at 72 °C for 10 min. PCR products were separated on 2% agarose gel by electrophoresis. 10% of samples from the study population were randomly selected for direct sequencing analysis to check the accuracy of genotyping.
Statistical analysisThe chi-squared (χ2) test was used to compute deviation from the Hardy-Weinberg Equilibrium (HWE) for two SNPs in the cases and controls. Genotypic and allelic frequencies were analyzed in each group using the χ2 and Fisher’s exact tests. T-test analyses were used to evaluate differences in the distribution of demographic variables. Conditional multivariate logistic regression analysis was used to calculate odds ratios (OR) and 95% confidence intervals (95% CI). Results were considered significant if the p-values were less than 0.05.
Analysis of the data was performed using SPSS v. 11.0 (SPSS, Inc. – Chicago, IL, United States).
ResultsIn total, 212 BD patients and 200 healthy controls were genotyped for the rs10515746 and rs9313439 SNPs. The allele and genotype frequencies for these SNPs are indicated in table 3.
Frequencies of alleles and genotypes of four TIM-3 SNPs in BD patients and controls
| SNPs | Alleles/ genotypes | BD n = 212 (%) | Controls n = 200 (%) | χ2 | p | OR (95% CI) |
|---|---|---|---|---|---|---|
| rs10515746 | GG | 55 (25.9) | 46 (23.2) | 0.408 | 0.96 | 1.03 (0.25-4.21) |
| Alleles | TG | 153 (72.2) | 148 (74.7) | 0.121 | 0.53 | 0.87 (0.55-1.36) |
| rs9313439 | TT | 4 (1.9) | 4 (2) | 0.492 | 0.73 | 0.95 (0.72-1.26) |
| Alleles | T | 162 (38.2) | 156 (39.4) | 0.177 | 0.50 | 1.19 (0.72-1.98) |
| G | 262 (61.8) | 240 (60.6) | 0.88 | 0.91 (0.27-3.05) | ||
| CC | 35 (16.5) | 38 (19.1) | 0.67 | 0.94 (0.71-1.24) | ||
| GC | 171(80.7) | 156 (78.4) | ||||
| GG | 6 (2.8) | 5 (2.5) | ||||
| C | 241(56.8) | 232 (58.3) | ||||
| G | 183(43.2) | 166(41.7) |
BD, Behçet disease; OD, odds ratio; 95% CI, 95% confidence interval.
Statistical analysis revealed that there were no significant differences between the BD patients (n = 212) and controls (n = 200) concerning the frequencies of rs10515746 and rs9313439. This study also did not indicate any associations between the SNPs and clinical presentations related to BD.
Of the 212 patients, 78 (36.8%) were women and 134 (63.2%) were men. The male to female ratio was 1.72 (134:78), and no statistically significant difference was observed in two genetic variants of TIM-3 between male and female patients. Moreover, when the study subjects were grouped according to age, (≤ 30 years and > 30 years), no significant correlation was detected between age groups and genotype frequency for any of the SNPs.
Hardy-Weinberg equilibrium (HWE)The rs10515746 and rs9313439 SNPs were not in HWE. In the authors’ opinion, departure from HWE was not considered to be due to typing errors, because part of the DNA samples was subjected to direct sequencing and all genotype results were concordant between the two methods (multiplex PCR and direct sequencing). Nonetheless, it is possible that some unidentified factors other than errors in genotyping data may explain deviation from HWE. For instance, the size of the sample or the impact of a complex mix of Iranian populations may have had an influence.
DiscussionThe role of dysregulated or abnormal immune responses in BD pathogenesis is well-defined and supports the theory that BD is an autoimmune disorder.15 The initiation and development of BD as an autoimmune disease is the result of a multistep pathogenic process, involving various genetic alterations and environmental factors leading to breakdown of tolerance and induction of an immune response against components of the self.
The transmembrane immunoglobulin and mucin domain (TIM) family comprises three genes in humans that encode the proteins TIM-1, TIM-3, and TIM-4. These molecules are expressed on different immune cell types and play important roles in the regulation of immune system.16,17 One of these three genes that could encode TIM-3 is specifically expressed by differentiated Th1 cells and acts as a negative regulator of Type 1 immunity.18 The proposed immunoregulatory role of TIM-3 has been strengthened using some animal models of Th1-mediated autoimmune diseases. For instance, anti-TIM-3 antibody exacerbated Th1-mediated experimental autoimmune encephalomyelitis (EAE) in an established animal model of multiple sclerosis.18 Similarly, TIM-3 pathway blockade in nonobese diabetic mice led to accelerated diabetes development, which have traditionally been viewed as a Th1-mediated pathology.19 Moreover, Zhu et al. indicated that administration of galectin-9, which is the best known ligand of TIM-3, causes the selective loss of interferon-gamma-producing cells and suppression of Th1 autoimmunity.20
Therefore, studying different aspects of this molecule is necessary, allowing a better understanding of the pathological mechanisms of autoimmunity, particularly in Th1-mediated autoimmune disorders, including BD.
Previous studies have reported increased TIM-3 gene expression in peripheral blood mononuclear cells of patients with BD compared with healthy controls. Moreover, a negative correlation between the frequency of TIM-3+ cells and BD activity has been observed in humans.21 Decreased expression of the TIM-3 ligand (galectin 9) in macrophages from BD-like mice has also been found compared with asymptomatic BD normal mice. Furthermore, injection of galactin 9 in BD-like mice resulted in the amelioration of inflammation, reduction in the severity score, and increased regulatory T cell expression.22 This evidence encouraged the authors to investigate the possible association of TIM-3 genetic polymorphisms with BD. To the authors’ knowledge, this is the first study relating TIM-3 gene polymorphisms with BD risk.
The results showed no association between TIM-3 SNPs and BD. In addition, there was no association between TIM-3 polymorphisms and BD after stratification by clinical features and gender.
The two SNPs selected in the present study have been previously shown to be associated with a variety of autoimmune diseases, including rheumatoid arthritis and multiple sclerosis.23,24
Nevertheless, none of these SNPs showed a significant association with BD. This is in agreement with previous observations that found no association between TIM-3 genetic polymorphisms and autoimmune conditions, including systemic lupus erythematosus, type 1 diabetes, and autoimmune thyroid diseases.14,25,26
Although the present study failed to show any association between the TIM-3 SNPs and BD, there is some evidence that the TIM-3 protein may play a role in the corresponding disease.21 There are several possible interpretations for these apparently different observations. One possible explanation could be related to the existence of different polymorphism loci in the TIM-3 gene that may influence risk of disease and were not tested in this study. The biological complexity of BD is another option. Because disease development can depend on both genetic and environmental factors, broad heterogeneity of the disease phenotype and predisposing factors may exist across different geographical regions. Therefore, caution must be used in the interpretation of results, and the association of TIM-3 polymorphisms with BD should be investigated using a larger sample size and other racial/ethnic populations.
ConclusionThis study failed to find any association between the two polymorphisms of TIM-3 gene (rs9313439 and rs10515746) and susceptibility for BD in Iranian patients. But due to the limited number of studies, its potential implications in disease pathogenesis must not be discarded and further studies are needed to evaluate the exact role of TIM-3 in BD.